3 key aspects regarding the interplay between the plant microbiome and its surrounding environments.
- Plant microbiome providing pathogen resistance
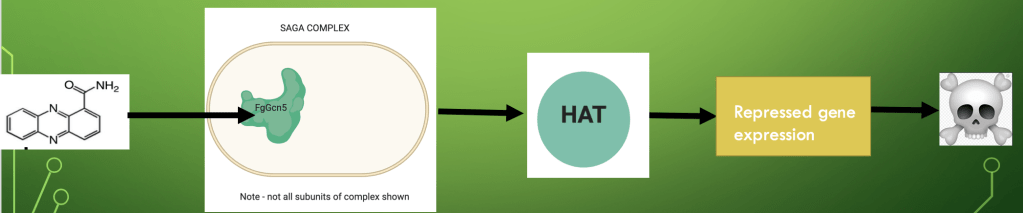
The plant microbiome can also be used to increase resistance to pathogens. As highlighted by (Chen et al., 2018) the ZJU60 bacteria isolated from the infected wheat head microbiome produced the secondary metabolite PCN which directly hindered the activity of the Fgcn5 protein in the SAGA complex of the fusarium gramarium fungus. This in turn resulted in inhibited histone acetyl transferase activity repressing gene expression and thus, fungal virulence of the pathogen. (Refer to figure 1) PCN produced by the bacteria was also shown by (Leland et al., 2010) to bring on induced system resistance in plants. Thus, elucidating how the interplay between the plant microbiome can increase pathogen resistance in it s host.
- Plant microbiome providing increased nutrients to host (nitrogen fixing)
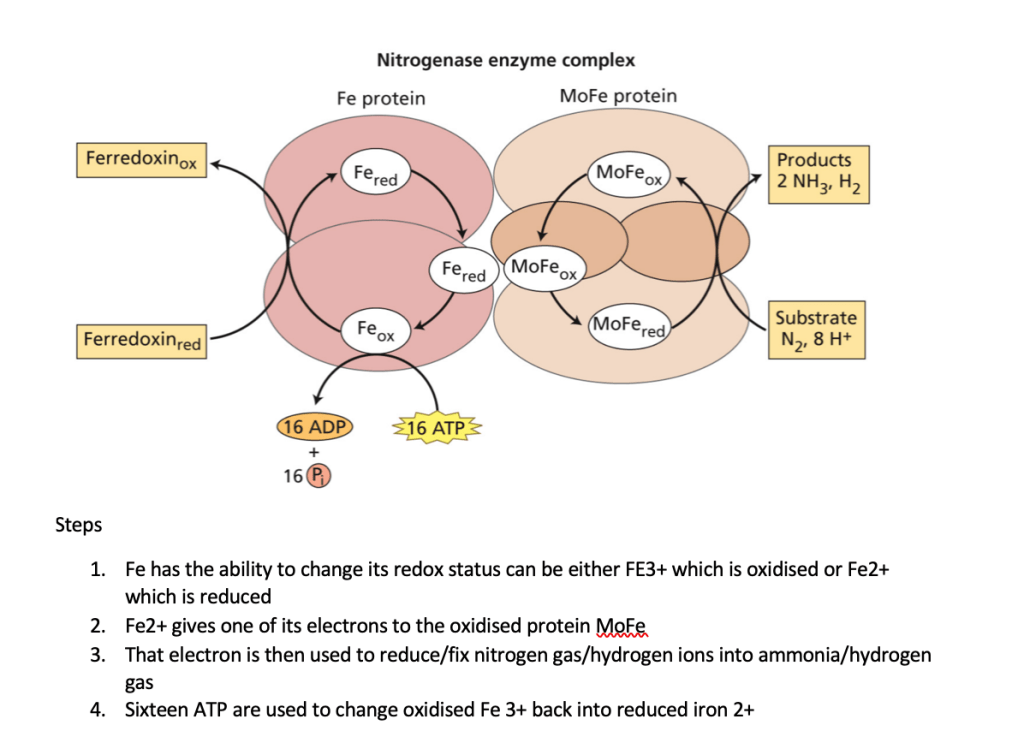
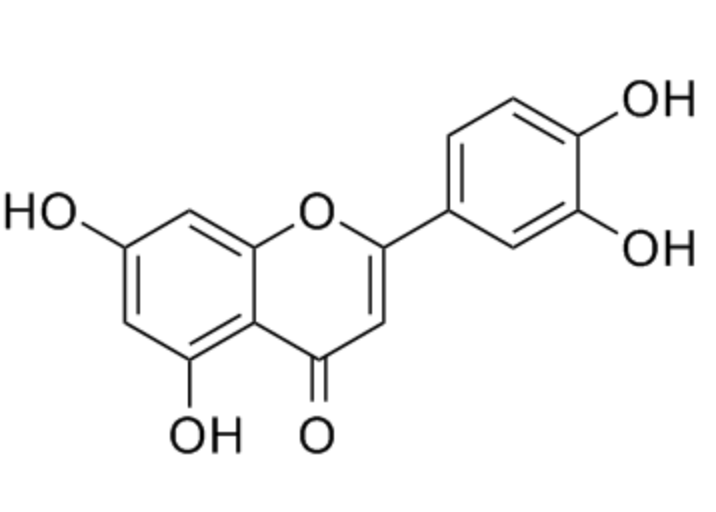
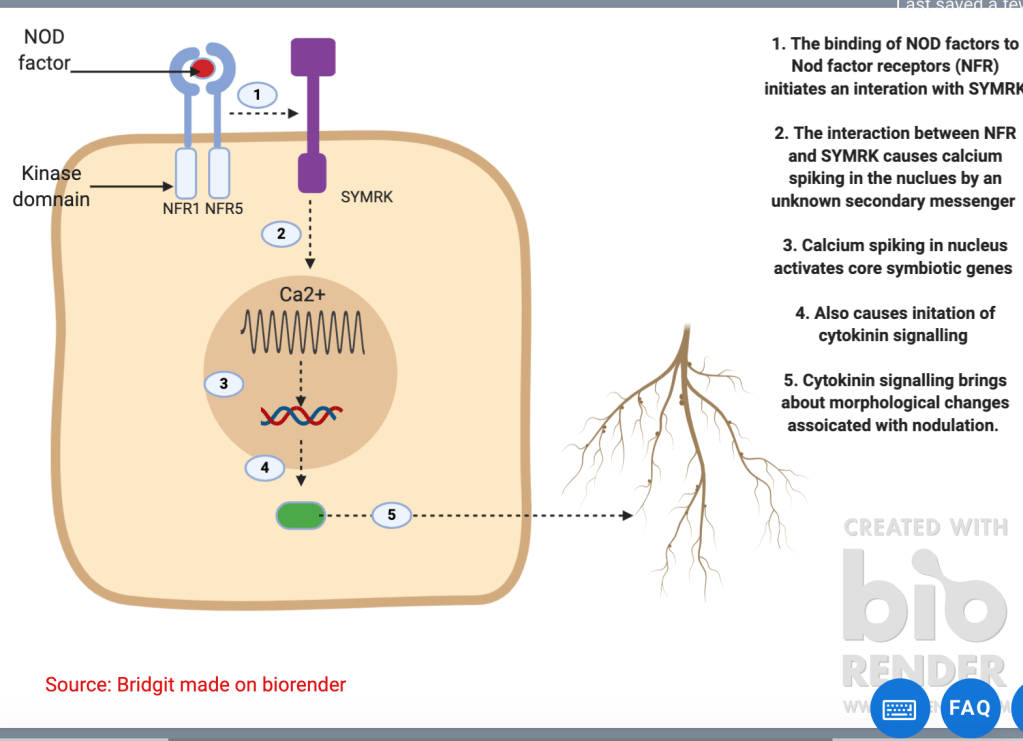
Microbes can increase nutrition uptake in plants. Rhizobia bacteria can fix atmospheric nitrogen N2 into ammonia – a form which can be utilised by plants. They do this via the nitrogenase enzyme complex, under anaerobic conditions. (Refer to figure 2). These bacteria can either be in the soil as free living rhizobia or within the root nodules of plants. To recruit bacteria into their roots plants secrete flavonoids such as luteolin (Figure 3). In response, bacteria produce Nod factors which are lipochitin oligosaccharides. These bind to Nod factor receptors at the plant cytoplasmic membrane. The interaction between NOD factor and the receptor results in protein phosphorylation. Protein phosphorylation activates symbiotic receptor like kinases (SYMRK) which causes calcium spiking in the nucleus. This signals the activation of calcium dependent protein kinases leading to the induction of expression of Nod genes required for nodule formation for the bacteria. In exchange for the nitrogen the plant will provide the Rhizobia bacteria with nutrition (primary sugars) and a protected environment to live in. Refer to figure 4 for illustration of this process.
- Plant microbiome providing stress relief to host
Plant growth-promoting rhizobacteria (PGPR) of the plant microbiome can increase plant growth under salt stress. The key PGPR involved in salt stress in plants are generally part of the endorhizosphere. They promote plant growth during salt stress through a number of ways. First, they directly provide the plant protective osmolytes including proline, trehalose and lysine betaines which can be directly up taken by plant roots. Protective osmolytes accumulate in the cytosol of cells in the plant to help maintain osmotic balance both within and outside the cell. They also help to prevent cellular oxidative damage under saline conditions. Second, they can increase the expression of genes associated with producing these protective osmolytes in the plant itself. For example, Bacillus megaterium from the Maize microbiome has been shown to also increase the expression of 14 genes known to increase salt tolerance in plants, this included the increased expression of p5CS1 which is involved in the production of the protective osmolyte proline. Third, PGPR including Anthrobacter contain ACC deminase which can break down excess ACC produced by plants during salt stress which lowers plant ethylene levels allowing IAA – the main type of auxin to promote plant growth.